
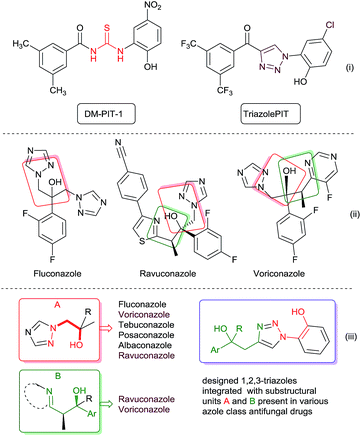
Pleckstrin, the protein where this domain was first detected, is the major substrate of protein kinase C in platelets.Proteins reported to contain PH domains belong to the following families: A Genome-wide look in Saccharomyces cerevisiae showed that most of the 33 yeast PH domains are indeed promiscuous in binding to phosphoinositides, while only one (Num1-PH) behaved highly specific. A large number of PH domains have poor affinity for phosphoinositides and are hypothesized to function as protein binding domains. Recruitment to the Golgi apparatus in this case is dependent on both PtdIns and ARF. PH domains can be found in many different proteins, such as OSBP or ARF. The only conserved residue among PH domains is a single tryptophan located within the alpha helix that serves to nucleate the core of the domain. The loops connecting the beta-strands differ greatly in length, making the PH domain relatively difficult to detect while providing the source of the domain's specificity. All known cases have a common structure consisting of two perpendicular anti-parallel beta sheets, followed by a C-terminal amphipathic helix. The 3D structure of several PH domains has been determined. Thus, such enzymes exert a part of their effect on cell function by modulating the localization of downstream signaling proteins that possess PH domains that are capable of binding their phospholipid products. This is important because it makes the recruitment of different PH domain containing proteins sensitive to the activities of enzymes that either phosphorylate or dephosphorylate these sites on the inositol ring, such as phosphoinositide 3-kinase or PTEN, respectively. Individual PH domains possess specificities for phosphoinositides phosphorylated at different sites within the inositol ring, e.g., some bind phosphatidylinositol (4,5)-bisphosphate but not phosphatidylinositol (3,4,5)-trisphosphate or phosphatidylinositol (3,4)-bisphosphate, while others may possess the requisite affinity. Through these interactions, PH domains play a role in recruiting proteins to different membranes, thus targeting them to appropriate cellular compartments or enabling them to interact with other components of the signal transduction pathways. This domain can bind phosphatidylinositol lipids within biological membranes (such as phosphatidylinositol (3,4,5)-trisphosphate and phosphatidylinositol (4,5)-bisphosphate), and proteins such as the βγ-subunits of heterotrimeric G proteins, and protein kinase C. Must be specified on their individual lines.Pleckstrin homology domain ( PH domain) or ( PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton.
Pip3 structure install#
The following options have an effect on the entire pip install run, and Requirements files only supports certain pip install options, which are listed Whitespaceįollowed by a # causes the # and the remainder of the line to beĬomments are stripped after line continuations are processed.
Comments #Ī line that begins with # is treated as a comment and ignored. Line continuations #Ī line ending in an unescaped \ is treated as a line continuationĪnd the newline following it is effectively ignored. PEP 263 style comments to change the encoding (i.e. Requirements files are utf-8 encoding by default and also support ForĮxamples of all these forms, see Examples. * python_version įor details on requirement specifiers, see Requirement Specifiers. pytest pytest - cov beautifulsoup4 # The syntax supported here is the same as that of requirement specifiers.

# It is possible to specify requirements as plain names. # This is a comment, to show how #-prefixed lines are ignored.


 0 kommentar(er)
0 kommentar(er)
